Templates
Atomik works with openEHR templates, which define each type of record or document that will be stored in Atomik. The template includes a data structure definition, data constraints and terminology, elements that Atomik uses to validate, process, store and query data created following those templates.
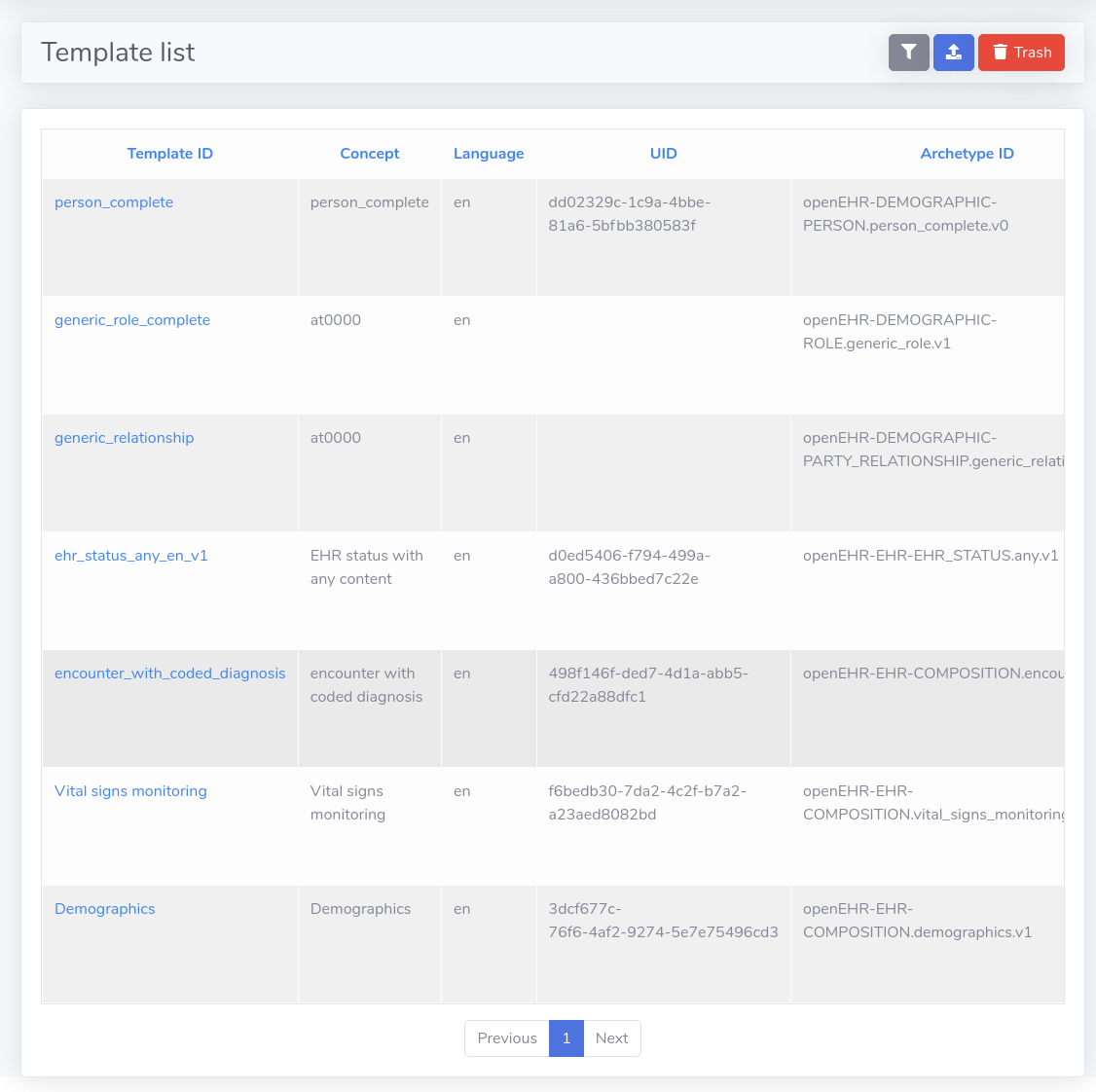
There are some templates publicly available in openEHR's Clinical Knowledge Manager, though templates are created mostly to satisfy local requirements. All openEHR templates are based on archetypes, which are general purpose definitions, and there are a lot of archetypes publicly available in the openEHR CKM. Of course, you can create your own archetype if you don't find what you need in the CKM.
There are many modeling tools available that allow you to create a template. We generally use Ocean's Template Designer.
Atomik has internal storage and management of templates, though you can use the openEHR Toolkit as a source of templates for Atomik. The Toolkit has some interesting tools that allow a better management of templates, and the use of templates to generate test data, that will help you to test while integrating Atomik into your platform.
Through the Web Console you can upload a template, which gets validated against the openEHR template XML schema (you will notice all templates are in XML format). Atomik indexes the templates (extracts pieces to be used for processing and querying data), and adds them to the in-memory cache, for quick access. Then when you create a query using the Query Builder, all the template information is used to define query conditions for filtering data.
Templates are versionable. That allows evolution of the data definitions in the system. Though old versions should still be available if there is existing data based on them. So querying old data used old template definitions, and querying new data uses new template definitions. If you model your templates carefully, maintaining backwards compatibility when possible, that will allow you to query old data by using the new templates. That is a recommendation we often do, so you can avoid some headaches in the future, because clinical data management and evolution is not a trivial task.