Getting Started
Welcome! This guide will get you up and running with Atomik in under 30 minutes. By the end, you'll store your first clinical record and retrieve it using our API.
Please let us know if you are interested in a specific example or guide we can build for this page.
What You'll Need
Before you begin:
- Atomik license credentials (DEV or PROD)
- Basic understanding of REST APIs
- HTTP client (curl, Postman, or your preferred tool)
- Optional: familiarity with openEHR concepts (we'll cover basics)
Quick Path Overview
Here's what we'll cover:
- Access your instance (5 min) - Get your endpoint and credentials
- Authenticate (5 min) - Make your first authenticated request
- Store data (10 min) - Commit your first clinical record
- Query data (10 min) - Retrieve what you just stored
Time required: ~30 minutes
Step 1: Access Your Atomik Instance
Once you receive your license, you'll get:- Your Atomik endpoint URL: https://your-instance.atomik.app
- API credentials
- Web Console access
Verify Your Connection
Test that you can reach your instance:
curl https://your-instance.atomik.appExpected response:
* Host https://your-instance.atomik.app was resolved.
* IPv4: 123.123.123.123
* Trying 123.123.123.123:443...
* Connected to your-instance.atomik.app (123.123.123.123) port 443
* TLSv1.3 (OUT), TLS handshake, Client hello (1):
* ...
* SSL connection using TLSv1.3 / TLS_AES_256_GCM_SHA384 / X25519 / id-ecPublicKey
* ALPN: server accepted http/1.1
* Server certificate:
* subject: CN=your-instance.atomik.app
* ...
* using HTTP/1.x
> GET / HTTP/1.1
> Host: your-instance.atomik.app
> User-Agent: curl/8.5.0
> Accept: */*✅ If you see this response, you're ready to proceed!
Step 2: Authentication
Atomik uses Bearer token authentication. First, obtain your access token:
curl --request POST \
--url https://your-instance.atomik.app/api/v1/auth \
--header 'Content-Type: application/x-www-form-urlencoded' \
--data email=your-username \
--data password=your-passwordResponse:
{
"token": "eyJhbGciOiJIUzI1Ni...Lc2ZtnR7ijjgma2qtLg"
}Test Authentication
Use your token to verify authentication works:
curl --request GET \
--url https://your-instance.atomik.app/api/v1/stats \
--header 'Authorization: Bearer eyJhbGciOiJIUzI1Ni...Lc2ZtnR7ijjgma2qtLg' \
✅ If you receive a successful response (even an empty list), authentication is working!
Step 3: Store Your First Clinical Record
Now let's store some clinical data. We'll use a simple blood pressure observation.
Create an EHR (Electronic Health Record)
First, create an EHR for a patient:
curl --request POST \
--url https://your-instance.atomik.app/api/v1/ehr \
--header 'Authorization: Bearer eyJhbGciOiJIUzI1Ni...Lc2ZtnR7ijjgma2qtLg' \
--header 'Prefer: return=representation' \
--header 'openEHR-AUDIT_DETAILS.committer: name="John Doe, MD", external_ref.id="BC8132EA-8F4A-11E7-BB31-BE2E44B06B35", external_ref.namespace="demographic", external_ref.type="PERSON"' \
--header 'openEHR-AUDIT_DETAILS.description: value="Created EHR by XYZ"' \Response:
{
"_type": "EHR",
"ehr_id": {
"_type": "HIER_OBJECT_ID",
"value": "d5384e04-eff4-45a4-94c8-c98ec17cdf42"
},
"system_id": {
"_type": "HIER_OBJECT_ID",
"value": "CABOLABS_ATOMIK_SERVER"
},
"time_created": {
"value": "2025-12-22 03:54:46"
},
"ehr_status": {
"_type": "EHR_STATUS",
"name": {
"_type": "DV_TEXT",
"value": "Generic Status"
},
"uid": {
"_type": "OBJECT_VERSION_ID",
"value": "3bdbed88-bb60-4f59-84ac-02abcd874dc5::CABOLABS_ATOMIK_SERVER::1"
},
"archetype_details": {
"archetype_id": {
"value": "openEHR-EHR-EHR_STATUS.generic.v1"
},
"template_id": {
"value": "ehr_status_generic_en_v1"
},
"rm_version": "1.0.2"
},
"archetype_node_id": "openEHR-EHR-EHR_STATUS.generic.v1",
"subject": {},
"is_modifiable": true,
"is_queryable": true
}
}💾 Save the ehr_id (d5384e04-eff4-45a4-94c8-c98ec17cdf42) - you'll need it for the next step!
Upload the openEHR template that defines the data:
Here you can find a sample template for storing just a Blood Pressure reading.
curl --request POST \
--url https://your-instance.atomik.app/api/v1/definition/template/adl1.4 \
--header 'Authorization: Bearer eyJhbGciOiJIUzI1Ni...Lc2ZtnR7ijjgma2qtLg' \
--header 'Content-Type: application/xml' \
--data '<?xml version="1.0" encoding="UTF-8" standalone="yes"?>
<template xmlns="http://schemas.openehr.org/v1">
<language>
<terminology_id>
<value>ISO_639-1</value>
</terminology_id>
<code_string>en</code_string>
</language>
<description>
<original_author id="date">2025-12-22</original_author>
<lifecycle_state>unmanaged</lifecycle_state>
<other_details id="licence"></other_details>
<other_details id="custodian_organisation"></other_details>
<other_details id="original_namespace"></other_details>
<other_details id="original_publisher"></other_details>
<other_details id="custodian_namespace"></other_details>
<other_details id="sem_ver">1.0.0</other_details>
<other_details id="build_uid">16c1f0c6-9fea-4469-a4cf-de92cdd4828f</other_details>
<details>
<language>
<terminology_id>
<value>ISO_639-1</value>
</terminology_id>
<code_string>en</code_string>
</language>
<purpose>Not Specified</purpose>
</details>
</description>
<uid>
<value>3212c78c-511e-46a5-a682-06dc4216fe80</value>
</uid>
<template_id>
<value>Blood Pressure</value>
</template_id>
<concept>Blood Pressure</concept>
<definition>
<rm_type_name>COMPOSITION</rm_type_name>
<occurrences>
<lower_included>true</lower_included>
<upper_included>true</upper_included>
<lower_unbounded>false</lower_unbounded>
<upper_unbounded>false</upper_unbounded>
<lower>1</lower>
<upper>1</upper>
</occurrences>
<node_id>at0000</node_id>
<attributes xsi:type="C_SINGLE_ATTRIBUTE" xmlns:xsi="http://www.w3.org/2001/XMLSchema-instance">
<rm_attribute_name>category</rm_attribute_name>
<existence>
<lower_included>true</lower_included>
<upper_included>true</upper_included>
<lower_unbounded>false</lower_unbounded>
<upper_unbounded>false</upper_unbounded>
<lower>1</lower>
<upper>1</upper>
</existence>
<children xsi:type="C_COMPLEX_OBJECT">
<rm_type_name>DV_CODED_TEXT</rm_type_name>
<occurrences>
<lower_included>true</lower_included>
<upper_included>true</upper_included>
<lower_unbounded>false</lower_unbounded>
<upper_unbounded>false</upper_unbounded>
<lower>1</lower>
<upper>1</upper>
</occurrences>
<node_id></node_id>
<attributes xsi:type="C_SINGLE_ATTRIBUTE">
<rm_attribute_name>defining_code</rm_attribute_name>
<existence>
<lower_included>true</lower_included>
<upper_included>true</upper_included>
<lower_unbounded>false</lower_unbounded>
<upper_unbounded>false</upper_unbounded>
<lower>1</lower>
<upper>1</upper>
</existence>
<children xsi:type="C_CODE_PHRASE">
<rm_type_name>CODE_PHRASE</rm_type_name>
<occurrences>
<lower_included>true</lower_included>
<upper_included>true</upper_included>
<lower_unbounded>false</lower_unbounded>
<upper_unbounded>false</upper_unbounded>
<lower>1</lower>
<upper>1</upper>
</occurrences>
<node_id></node_id>
<terminology_id>
<value>openehr</value>
</terminology_id>
<code_list>433</code_list>
</children>
</attributes>
</children>
</attributes>
<attributes xsi:type="C_SINGLE_ATTRIBUTE" xmlns:xsi="http://www.w3.org/2001/XMLSchema-instance">
<rm_attribute_name>context</rm_attribute_name>
<existence>
<lower_included>true</lower_included>
<upper_included>true</upper_included>
<lower_unbounded>false</lower_unbounded>
<upper_unbounded>false</upper_unbounded>
<lower>0</lower>
<upper>1</upper>
</existence>
<children xsi:type="C_COMPLEX_OBJECT">
<rm_type_name>EVENT_CONTEXT</rm_type_name>
<occurrences>
<lower_included>true</lower_included>
<upper_included>true</upper_included>
<lower_unbounded>false</lower_unbounded>
<upper_unbounded>false</upper_unbounded>
<lower>1</lower>
<upper>1</upper>
</occurrences>
<node_id></node_id>
<attributes xsi:type="C_SINGLE_ATTRIBUTE">
<rm_attribute_name>other_context</rm_attribute_name>
<existence>
<lower_included>true</lower_included>
<upper_included>true</upper_included>
<lower_unbounded>false</lower_unbounded>
<upper_unbounded>false</upper_unbounded>
<lower>0</lower>
<upper>1</upper>
</existence>
<children xsi:type="C_COMPLEX_OBJECT">
<rm_type_name>ITEM_TREE</rm_type_name>
<occurrences>
<lower_included>true</lower_included>
<upper_included>true</upper_included>
<lower_unbounded>false</lower_unbounded>
<upper_unbounded>false</upper_unbounded>
<lower>1</lower>
<upper>1</upper>
</occurrences>
<node_id>at0001</node_id>
<attributes xsi:type="C_MULTIPLE_ATTRIBUTE">
<rm_attribute_name>items</rm_attribute_name>
<existence>
<lower_included>true</lower_included>
<upper_included>true</upper_included>
<lower_unbounded>false</lower_unbounded>
<upper_unbounded>false</upper_unbounded>
<lower>0</lower>
<upper>1</upper>
</existence>
<children xsi:type="ARCHETYPE_SLOT">
<rm_type_name>CLUSTER</rm_type_name>
<occurrences>
<lower_included>true</lower_included>
<upper_included>false</upper_included>
<lower_unbounded>false</lower_unbounded>
<upper_unbounded>true</upper_unbounded>
<lower>0</lower>
</occurrences>
<node_id>at0002</node_id>
<includes>
<string_expression>archetype_id/value matches {/.*/}</string_expression>
<expression xsi:type="EXPR_BINARY_OPERATOR">
<type>Boolean</type>
<operator>2007</operator>
<precedence_overridden>false</precedence_overridden>
<left_operand xsi:type="EXPR_LEAF">
<type>String</type>
<item xsi:type="xs:string" xmlns:xs="http://www.w3.org/2001/XMLSchema">archetype_id/value</item>
<reference_type>attribute</reference_type>
</left_operand>
<right_operand xsi:type="EXPR_LEAF">
<type>String</type>
<item xsi:type="C_STRING">
<pattern>.*</pattern>
</item>
<reference_type>constraint</reference_type>
</right_operand>
</expression>
</includes>
</children>
<cardinality>
<is_ordered>false</is_ordered>
<is_unique>false</is_unique>
<interval>
<lower_included>true</lower_included>
<upper_included>false</upper_included>
<lower_unbounded>false</lower_unbounded>
<upper_unbounded>true</upper_unbounded>
<lower>0</lower>
</interval>
</cardinality>
</attributes>
</children>
</attributes>
</children>
</attributes>
<attributes xsi:type="C_MULTIPLE_ATTRIBUTE" xmlns:xsi="http://www.w3.org/2001/XMLSchema-instance">
<rm_attribute_name>content</rm_attribute_name>
<existence>
<lower_included>true</lower_included>
<upper_included>true</upper_included>
<lower_unbounded>false</lower_unbounded>
<upper_unbounded>false</upper_unbounded>
<lower>0</lower>
<upper>1</upper>
</existence>
<children xsi:type="C_ARCHETYPE_ROOT">
<rm_type_name>OBSERVATION</rm_type_name>
<occurrences>
<lower_included>true</lower_included>
<upper_included>true</upper_included>
<lower_unbounded>false</lower_unbounded>
<upper_unbounded>false</upper_unbounded>
<lower>0</lower>
<upper>1</upper>
</occurrences>
<node_id>at0000</node_id>
<attributes xsi:type="C_SINGLE_ATTRIBUTE">
<rm_attribute_name>data</rm_attribute_name>
<existence>
<lower_included>true</lower_included>
<upper_included>true</upper_included>
<lower_unbounded>false</lower_unbounded>
<upper_unbounded>false</upper_unbounded>
<lower>1</lower>
<upper>1</upper>
</existence>
<children xsi:type="C_COMPLEX_OBJECT">
<rm_type_name>HISTORY</rm_type_name>
<occurrences>
<lower_included>true</lower_included>
<upper_included>true</upper_included>
<lower_unbounded>false</lower_unbounded>
<upper_unbounded>false</upper_unbounded>
<lower>1</lower>
<upper>1</upper>
</occurrences>
<node_id>at0001</node_id>
<attributes xsi:type="C_MULTIPLE_ATTRIBUTE">
<rm_attribute_name>events</rm_attribute_name>
<existence>
<lower_included>true</lower_included>
<upper_included>true</upper_included>
<lower_unbounded>false</lower_unbounded>
<upper_unbounded>false</upper_unbounded>
<lower>0</lower>
<upper>1</upper>
</existence>
<children xsi:type="C_COMPLEX_OBJECT">
<rm_type_name>EVENT</rm_type_name>
<occurrences>
<lower_included>true</lower_included>
<upper_included>false</upper_included>
<lower_unbounded>false</lower_unbounded>
<upper_unbounded>true</upper_unbounded>
<lower>0</lower>
</occurrences>
<node_id>at0006</node_id>
<attributes xsi:type="C_SINGLE_ATTRIBUTE">
<rm_attribute_name>data</rm_attribute_name>
<existence>
<lower_included>true</lower_included>
<upper_included>true</upper_included>
<lower_unbounded>false</lower_unbounded>
<upper_unbounded>false</upper_unbounded>
<lower>1</lower>
<upper>1</upper>
</existence>
<children xsi:type="C_COMPLEX_OBJECT">
<rm_type_name>ITEM_TREE</rm_type_name>
<occurrences>
<lower_included>true</lower_included>
<upper_included>true</upper_included>
<lower_unbounded>false</lower_unbounded>
<upper_unbounded>false</upper_unbounded>
<lower>1</lower>
<upper>1</upper>
</occurrences>
<node_id>at0003</node_id>
<attributes xsi:type="C_MULTIPLE_ATTRIBUTE">
<rm_attribute_name>items</rm_attribute_name>
<existence>
<lower_included>true</lower_included>
<upper_included>true</upper_included>
<lower_unbounded>false</lower_unbounded>
<upper_unbounded>false</upper_unbounded>
<lower>0</lower>
<upper>1</upper>
</existence>
<children xsi:type="C_COMPLEX_OBJECT">
<rm_type_name>ELEMENT</rm_type_name>
<occurrences>
<lower_included>true</lower_included>
<upper_included>true</upper_included>
<lower_unbounded>false</lower_unbounded>
<upper_unbounded>false</upper_unbounded>
<lower>0</lower>
<upper>1</upper>
</occurrences>
<node_id>at0004</node_id>
<attributes xsi:type="C_SINGLE_ATTRIBUTE">
<rm_attribute_name>value</rm_attribute_name>
<existence>
<lower_included>true</lower_included>
<upper_included>true</upper_included>
<lower_unbounded>false</lower_unbounded>
<upper_unbounded>false</upper_unbounded>
<lower>0</lower>
<upper>1</upper>
</existence>
<children xsi:type="C_DV_QUANTITY">
<rm_type_name>DV_QUANTITY</rm_type_name>
<occurrences>
<lower_included>true</lower_included>
<upper_included>true</upper_included>
<lower_unbounded>false</lower_unbounded>
<upper_unbounded>false</upper_unbounded>
<lower>1</lower>
<upper>1</upper>
</occurrences>
<node_id></node_id>
<property>
<terminology_id>
<value>openehr</value>
</terminology_id>
<code_string>125</code_string>
</property>
<list>
<magnitude>
<lower_included>true</lower_included>
<upper_included>false</upper_included>
<lower_unbounded>false</lower_unbounded>
<upper_unbounded>false</upper_unbounded>
<lower>0.0</lower>
<upper>1000.0</upper>
</magnitude>
<precision>
<lower_included>true</lower_included>
<upper_included>true</upper_included>
<lower_unbounded>false</lower_unbounded>
<upper_unbounded>false</upper_unbounded>
<lower>0</lower>
<upper>0</upper>
</precision>
<units>mm[Hg]</units>
</list>
</children>
</attributes>
</children>
<children xsi:type="C_COMPLEX_OBJECT">
<rm_type_name>ELEMENT</rm_type_name>
<occurrences>
<lower_included>true</lower_included>
<upper_included>true</upper_included>
<lower_unbounded>false</lower_unbounded>
<upper_unbounded>false</upper_unbounded>
<lower>0</lower>
<upper>1</upper>
</occurrences>
<node_id>at0005</node_id>
<attributes xsi:type="C_SINGLE_ATTRIBUTE">
<rm_attribute_name>value</rm_attribute_name>
<existence>
<lower_included>true</lower_included>
<upper_included>true</upper_included>
<lower_unbounded>false</lower_unbounded>
<upper_unbounded>false</upper_unbounded>
<lower>0</lower>
<upper>1</upper>
</existence>
<children xsi:type="C_DV_QUANTITY">
<rm_type_name>DV_QUANTITY</rm_type_name>
<occurrences>
<lower_included>true</lower_included>
<upper_included>true</upper_included>
<lower_unbounded>false</lower_unbounded>
<upper_unbounded>false</upper_unbounded>
<lower>1</lower>
<upper>1</upper>
</occurrences>
<node_id></node_id>
<property>
<terminology_id>
<value>openehr</value>
</terminology_id>
<code_string>125</code_string>
</property>
<list>
<magnitude>
<lower_included>true</lower_included>
<upper_included>false</upper_included>
<lower_unbounded>false</lower_unbounded>
<upper_unbounded>false</upper_unbounded>
<lower>0.0</lower>
<upper>1000.0</upper>
</magnitude>
<precision>
<lower_included>true</lower_included>
<upper_included>true</upper_included>
<lower_unbounded>false</lower_unbounded>
<upper_unbounded>false</upper_unbounded>
<lower>0</lower>
<upper>0</upper>
</precision>
<units>mm[Hg]</units>
</list>
</children>
</attributes>
</children>
<children xsi:type="C_COMPLEX_OBJECT">
<rm_type_name>ELEMENT</rm_type_name>
<occurrences>
<lower_included>true</lower_included>
<upper_included>true</upper_included>
<lower_unbounded>false</lower_unbounded>
<upper_unbounded>false</upper_unbounded>
<lower>0</lower>
<upper>1</upper>
</occurrences>
<node_id>at1007</node_id>
<attributes xsi:type="C_SINGLE_ATTRIBUTE">
<rm_attribute_name>value</rm_attribute_name>
<existence>
<lower_included>true</lower_included>
<upper_included>true</upper_included>
<lower_unbounded>false</lower_unbounded>
<upper_unbounded>false</upper_unbounded>
<lower>0</lower>
<upper>1</upper>
</existence>
<children xsi:type="C_DV_QUANTITY">
<rm_type_name>DV_QUANTITY</rm_type_name>
<occurrences>
<lower_included>true</lower_included>
<upper_included>true</upper_included>
<lower_unbounded>false</lower_unbounded>
<upper_unbounded>false</upper_unbounded>
<lower>1</lower>
<upper>1</upper>
</occurrences>
<node_id></node_id>
<property>
<terminology_id>
<value>openehr</value>
</terminology_id>
<code_string>125</code_string>
</property>
<list>
<magnitude>
<lower_included>true</lower_included>
<upper_included>false</upper_included>
<lower_unbounded>false</lower_unbounded>
<upper_unbounded>false</upper_unbounded>
<lower>0.0</lower>
<upper>1000.0</upper>
</magnitude>
<precision>
<lower_included>true</lower_included>
<upper_included>true</upper_included>
<lower_unbounded>false</lower_unbounded>
<upper_unbounded>false</upper_unbounded>
<lower>0</lower>
<upper>0</upper>
</precision>
<units>mm[Hg]</units>
</list>
</children>
</attributes>
</children>
<children xsi:type="C_COMPLEX_OBJECT">
<rm_type_name>ELEMENT</rm_type_name>
<occurrences>
<lower_included>true</lower_included>
<upper_included>true</upper_included>
<lower_unbounded>false</lower_unbounded>
<upper_unbounded>false</upper_unbounded>
<lower>0</lower>
<upper>1</upper>
</occurrences>
<node_id>at1059</node_id>
<attributes xsi:type="C_SINGLE_ATTRIBUTE">
<rm_attribute_name>value</rm_attribute_name>
<existence>
<lower_included>true</lower_included>
<upper_included>true</upper_included>
<lower_unbounded>false</lower_unbounded>
<upper_unbounded>false</upper_unbounded>
<lower>0</lower>
<upper>1</upper>
</existence>
<children xsi:type="C_COMPLEX_OBJECT">
<rm_type_name>DV_TEXT</rm_type_name>
<occurrences>
<lower_included>true</lower_included>
<upper_included>true</upper_included>
<lower_unbounded>false</lower_unbounded>
<upper_unbounded>false</upper_unbounded>
<lower>1</lower>
<upper>1</upper>
</occurrences>
<node_id></node_id>
</children>
</attributes>
</children>
<children xsi:type="C_COMPLEX_OBJECT">
<rm_type_name>ELEMENT</rm_type_name>
<occurrences>
<lower_included>true</lower_included>
<upper_included>true</upper_included>
<lower_unbounded>false</lower_unbounded>
<upper_unbounded>false</upper_unbounded>
<lower>0</lower>
<upper>1</upper>
</occurrences>
<node_id>at0033</node_id>
<attributes xsi:type="C_SINGLE_ATTRIBUTE">
<rm_attribute_name>value</rm_attribute_name>
<existence>
<lower_included>true</lower_included>
<upper_included>true</upper_included>
<lower_unbounded>false</lower_unbounded>
<upper_unbounded>false</upper_unbounded>
<lower>0</lower>
<upper>1</upper>
</existence>
<children xsi:type="C_COMPLEX_OBJECT">
<rm_type_name>DV_TEXT</rm_type_name>
<occurrences>
<lower_included>true</lower_included>
<upper_included>true</upper_included>
<lower_unbounded>false</lower_unbounded>
<upper_unbounded>false</upper_unbounded>
<lower>1</lower>
<upper>1</upper>
</occurrences>
<node_id></node_id>
</children>
</attributes>
</children>
<cardinality>
<is_ordered>false</is_ordered>
<is_unique>false</is_unique>
<interval>
<lower_included>true</lower_included>
<upper_included>false</upper_included>
<lower_unbounded>false</lower_unbounded>
<upper_unbounded>true</upper_unbounded>
<lower>0</lower>
</interval>
</cardinality>
</attributes>
</children>
</attributes>
<attributes xsi:type="C_SINGLE_ATTRIBUTE">
<rm_attribute_name>state</rm_attribute_name>
<existence>
<lower_included>true</lower_included>
<upper_included>true</upper_included>
<lower_unbounded>false</lower_unbounded>
<upper_unbounded>false</upper_unbounded>
<lower>0</lower>
<upper>1</upper>
</existence>
<children xsi:type="C_COMPLEX_OBJECT">
<rm_type_name>ITEM_TREE</rm_type_name>
<occurrences>
<lower_included>true</lower_included>
<upper_included>true</upper_included>
<lower_unbounded>false</lower_unbounded>
<upper_unbounded>false</upper_unbounded>
<lower>1</lower>
<upper>1</upper>
</occurrences>
<node_id>at0007</node_id>
<attributes xsi:type="C_MULTIPLE_ATTRIBUTE">
<rm_attribute_name>items</rm_attribute_name>
<existence>
<lower_included>true</lower_included>
<upper_included>true</upper_included>
<lower_unbounded>false</lower_unbounded>
<upper_unbounded>false</upper_unbounded>
<lower>0</lower>
<upper>1</upper>
</existence>
<children xsi:type="C_COMPLEX_OBJECT">
<rm_type_name>ELEMENT</rm_type_name>
<occurrences>
<lower_included>true</lower_included>
<upper_included>true</upper_included>
<lower_unbounded>false</lower_unbounded>
<upper_unbounded>false</upper_unbounded>
<lower>0</lower>
<upper>1</upper>
</occurrences>
<node_id>at0008</node_id>
<attributes xsi:type="C_SINGLE_ATTRIBUTE">
<rm_attribute_name>value</rm_attribute_name>
<existence>
<lower_included>true</lower_included>
<upper_included>true</upper_included>
<lower_unbounded>false</lower_unbounded>
<upper_unbounded>false</upper_unbounded>
<lower>0</lower>
<upper>1</upper>
</existence>
<children xsi:type="C_COMPLEX_OBJECT">
<rm_type_name>DV_CODED_TEXT</rm_type_name>
<occurrences>
<lower_included>true</lower_included>
<upper_included>true</upper_included>
<lower_unbounded>false</lower_unbounded>
<upper_unbounded>false</upper_unbounded>
<lower>1</lower>
<upper>1</upper>
</occurrences>
<node_id></node_id>
<attributes xsi:type="C_SINGLE_ATTRIBUTE">
<rm_attribute_name>defining_code</rm_attribute_name>
<existence>
<lower_included>true</lower_included>
<upper_included>true</upper_included>
<lower_unbounded>false</lower_unbounded>
<upper_unbounded>false</upper_unbounded>
<lower>1</lower>
<upper>1</upper>
</existence>
<children xsi:type="C_CODE_PHRASE">
<rm_type_name>CODE_PHRASE</rm_type_name>
<occurrences>
<lower_included>true</lower_included>
<upper_included>true</upper_included>
<lower_unbounded>false</lower_unbounded>
<upper_unbounded>false</upper_unbounded>
<lower>1</lower>
<upper>1</upper>
</occurrences>
<node_id></node_id>
<terminology_id>
<value>local</value>
</terminology_id>
<code_list>at1000</code_list>
<code_list>at1001</code_list>
<code_list>at1002</code_list>
<code_list>at1003</code_list>
<code_list>at1014</code_list>
</children>
</attributes>
</children>
</attributes>
</children>
<children xsi:type="ARCHETYPE_SLOT">
<rm_type_name>CLUSTER</rm_type_name>
<occurrences>
<lower_included>true</lower_included>
<upper_included>true</upper_included>
<lower_unbounded>false</lower_unbounded>
<upper_unbounded>false</upper_unbounded>
<lower>0</lower>
<upper>1</upper>
</occurrences>
<node_id>at1030</node_id>
<includes>
<string_expression>archetype_id/value matches {/openEHR-EHR-CLUSTER\.level_of_exertion(-[a-zA-Z0-9_]+)*\.v1/}</string_expression>
<expression xsi:type="EXPR_BINARY_OPERATOR">
<type>Boolean</type>
<operator>2007</operator>
<precedence_overridden>false</precedence_overridden>
<left_operand xsi:type="EXPR_LEAF">
<type>String</type>
<item xsi:type="xs:string" xmlns:xs="http://www.w3.org/2001/XMLSchema">archetype_id/value</item>
<reference_type>attribute</reference_type>
</left_operand>
<right_operand xsi:type="EXPR_LEAF">
<type>String</type>
<item xsi:type="C_STRING">
<pattern>openEHR-EHR-CLUSTER\.level_of_exertion(-[a-zA-Z0-9_]+)*\.v1</pattern>
</item>
<reference_type>constraint</reference_type>
</right_operand>
</expression>
</includes>
</children>
<cardinality>
<is_ordered>false</is_ordered>
<is_unique>false</is_unique>
<interval>
<lower_included>true</lower_included>
<upper_included>false</upper_included>
<lower_unbounded>false</lower_unbounded>
<upper_unbounded>true</upper_unbounded>
<lower>0</lower>
</interval>
</cardinality>
</attributes>
</children>
</attributes>
</children>
<cardinality>
<is_ordered>false</is_ordered>
<is_unique>false</is_unique>
<interval>
<lower_included>true</lower_included>
<upper_included>false</upper_included>
<lower_unbounded>false</lower_unbounded>
<upper_unbounded>true</upper_unbounded>
<lower>1</lower>
</interval>
</cardinality>
</attributes>
</children>
</attributes>
<attributes xsi:type="C_SINGLE_ATTRIBUTE">
<rm_attribute_name>protocol</rm_attribute_name>
<existence>
<lower_included>true</lower_included>
<upper_included>true</upper_included>
<lower_unbounded>false</lower_unbounded>
<upper_unbounded>false</upper_unbounded>
<lower>0</lower>
<upper>1</upper>
</existence>
<children xsi:type="C_COMPLEX_OBJECT">
<rm_type_name>ITEM_TREE</rm_type_name>
<occurrences>
<lower_included>true</lower_included>
<upper_included>true</upper_included>
<lower_unbounded>false</lower_unbounded>
<upper_unbounded>false</upper_unbounded>
<lower>1</lower>
<upper>1</upper>
</occurrences>
<node_id>at0011</node_id>
<attributes xsi:type="C_MULTIPLE_ATTRIBUTE">
<rm_attribute_name>items</rm_attribute_name>
<existence>
<lower_included>true</lower_included>
<upper_included>true</upper_included>
<lower_unbounded>false</lower_unbounded>
<upper_unbounded>false</upper_unbounded>
<lower>0</lower>
<upper>1</upper>
</existence>
<children xsi:type="ARCHETYPE_SLOT">
<rm_type_name>CLUSTER</rm_type_name>
<occurrences>
<lower_included>true</lower_included>
<upper_included>false</upper_included>
<lower_unbounded>false</lower_unbounded>
<upper_unbounded>true</upper_unbounded>
<lower>0</lower>
</occurrences>
<node_id>at1057</node_id>
<includes>
<string_expression>archetype_id/value matches {/openEHR-EHR-CLUSTER\.anatomical_location(-[a-zA-Z0-9_]+)*\.v1/}</string_expression>
<expression xsi:type="EXPR_BINARY_OPERATOR">
<type>Boolean</type>
<operator>2007</operator>
<precedence_overridden>false</precedence_overridden>
<left_operand xsi:type="EXPR_LEAF">
<type>String</type>
<item xsi:type="xs:string" xmlns:xs="http://www.w3.org/2001/XMLSchema">archetype_id/value</item>
<reference_type>attribute</reference_type>
</left_operand>
<right_operand xsi:type="EXPR_LEAF">
<type>String</type>
<item xsi:type="C_STRING">
<pattern>openEHR-EHR-CLUSTER\.anatomical_location(-[a-zA-Z0-9_]+)*\.v1</pattern>
</item>
<reference_type>constraint</reference_type>
</right_operand>
</expression>
</includes>
</children>
<children xsi:type="ARCHETYPE_SLOT">
<rm_type_name>CLUSTER</rm_type_name>
<occurrences>
<lower_included>true</lower_included>
<upper_included>true</upper_included>
<lower_unbounded>false</lower_unbounded>
<upper_unbounded>false</upper_unbounded>
<lower>0</lower>
<upper>1</upper>
</occurrences>
<node_id>at1025</node_id>
<includes>
<string_expression>archetype_id/value matches {/openEHR-EHR-CLUSTER\.device(-[a-zA-Z0-9_]+)*\.v1/}</string_expression>
<expression xsi:type="EXPR_BINARY_OPERATOR">
<type>Boolean</type>
<operator>2007</operator>
<precedence_overridden>false</precedence_overridden>
<left_operand xsi:type="EXPR_LEAF">
<type>String</type>
<item xsi:type="xs:string" xmlns:xs="http://www.w3.org/2001/XMLSchema">archetype_id/value</item>
<reference_type>attribute</reference_type>
</left_operand>
<right_operand xsi:type="EXPR_LEAF">
<type>String</type>
<item xsi:type="C_STRING">
<pattern>openEHR-EHR-CLUSTER\.device(-[a-zA-Z0-9_]+)*\.v1</pattern>
</item>
<reference_type>constraint</reference_type>
</right_operand>
</expression>
</includes>
</children>
<children xsi:type="ARCHETYPE_SLOT">
<rm_type_name>CLUSTER</rm_type_name>
<occurrences>
<lower_included>true</lower_included>
<upper_included>false</upper_included>
<lower_unbounded>false</lower_unbounded>
<upper_unbounded>true</upper_unbounded>
<lower>0</lower>
</occurrences>
<node_id>at1058</node_id>
<includes>
<string_expression>archetype_id/value matches {/.*/}</string_expression>
<expression xsi:type="EXPR_BINARY_OPERATOR">
<type>Boolean</type>
<operator>2007</operator>
<precedence_overridden>false</precedence_overridden>
<left_operand xsi:type="EXPR_LEAF">
<type>String</type>
<item xsi:type="xs:string" xmlns:xs="http://www.w3.org/2001/XMLSchema">archetype_id/value</item>
<reference_type>attribute</reference_type>
</left_operand>
<right_operand xsi:type="EXPR_LEAF">
<type>String</type>
<item xsi:type="C_STRING">
<pattern>.*</pattern>
</item>
<reference_type>constraint</reference_type>
</right_operand>
</expression>
</includes>
</children>
<cardinality>
<is_ordered>false</is_ordered>
<is_unique>false</is_unique>
<interval>
<lower_included>true</lower_included>
<upper_included>false</upper_included>
<lower_unbounded>false</lower_unbounded>
<upper_unbounded>true</upper_unbounded>
<lower>0</lower>
</interval>
</cardinality>
</attributes>
</children>
</attributes>
<archetype_id>
<value>openEHR-EHR-OBSERVATION.blood_pressure.v2</value>
</archetype_id>
<term_definitions code="at0001">
<items id="text">History</items>
<items id="description">History Structural node.</items>
</term_definitions>
<term_definitions code="at0003">
<items id="text">blood pressure</items>
<items id="description">@ internal @</items>
</term_definitions>
<term_definitions code="at0004">
<items id="text">Systolic</items>
<items id="description">Peak systemic arterial blood pressure - measured in systolic or contraction phase of the heart cycle.</items>
</term_definitions>
<term_definitions code="at0005">
<items id="text">Diastolic</items>
<items id="description">Minimum systemic arterial blood pressure - measured in the diastolic or relaxation phase of the heart cycle.</items>
</term_definitions>
<term_definitions code="at0006">
<items id="text">Any event</items>
<items id="description">Default, unspecified point in time or interval event which may be explicitly defined in a template or at run-time.</items>
</term_definitions>
<term_definitions code="at0007">
<items id="text">state structure</items>
<items id="description">@ internal @</items>
</term_definitions>
<term_definitions code="at0008">
<items id="text">Position</items>
<items id="description">The position of the individual at the time of measurement.</items>
</term_definitions>
<term_definitions code="at0011">
<items id="text">Tree</items>
<items id="description">List structure.</items>
</term_definitions>
<term_definitions code="at0013">
<items id="text">Cuff size</items>
<items id="description">The size of the cuff used for blood pressure measurement.</items>
<items id="comment">Perloff D, Grim C, Flack J, Frohlich ED, Hill M, McDonald M, Morgenstern BZ. Human blood pressure determination by sphygmomanometry. Circulation 1993;88;2460-2470. </items>
</term_definitions>
<term_definitions code="at0014">
<items id="text">Location of measurement</items>
<items id="description">Simple body site where blood pressure was measured.</items>
</term_definitions>
<term_definitions code="at0015">
<items id="text">Adult Thigh</items>
<items id="description">A cuff used for an adult thigh.</items>
</term_definitions>
<term_definitions code="at0016">
<items id="text">Large Adult</items>
<items id="description">A cuff for adults with larger arms.</items>
</term_definitions>
<term_definitions code="at0017">
<items id="text">Adult</items>
<items id="description">A cuff that is standard for an adult.</items>
</term_definitions>
<term_definitions code="at0025">
<items id="text">Right arm</items>
<items id="description">The right arm of the person.</items>
</term_definitions>
<term_definitions code="at0026">
<items id="text">Left arm</items>
<items id="description">The left arm of the person.</items>
</term_definitions>
<term_definitions code="at0027">
<items id="text">Right thigh</items>
<items id="description">The right thigh of the person.</items>
</term_definitions>
<term_definitions code="at0028">
<items id="text">Left thigh</items>
<items id="description">The left thigh of the person.</items>
</term_definitions>
<term_definitions code="at0033">
<items id="text">Comment</items>
<items id="description">Additional narrative about the measurement, not captured in other fields.</items>
</term_definitions>
<term_definitions code="at1000">
<items id="text">Standing</items>
<items id="description">Standing at the time of blood pressure measurement.</items>
</term_definitions>
<term_definitions code="at1001">
<items id="text">Sitting</items>
<items id="description">Sitting (for example on bed or chair) at the time of blood pressure measurement.</items>
</term_definitions>
<term_definitions code="at1002">
<items id="text">Reclining</items>
<items id="description">Reclining at the time of blood pressure measurement.</items>
</term_definitions>
<term_definitions code="at1003">
<items id="text">Lying</items>
<items id="description">Lying flat at the time of blood pressure measurement.</items>
</term_definitions>
<term_definitions code="at1005">
<items id="text">Tilt</items>
<items id="description">The craniocaudal tilt of the surface on which the person is lying at the time of measurement.</items>
</term_definitions>
<term_definitions code="at1006">
<items id="text">Mean arterial pressure</items>
<items id="description">The average arterial pressure that occurs over the entire course of the heart contraction and relaxation cycle.</items>
</term_definitions>
<term_definitions code="at1007">
<items id="text">Pulse pressure</items>
<items id="description">The difference between the systolic and diastolic pressure.</items>
</term_definitions>
<term_definitions code="at1008">
<items id="text">Small Adult</items>
<items id="description">A cuff used for a small adult.</items>
</term_definitions>
<term_definitions code="at1009">
<items id="text">Paediatric/Child</items>
<items id="description">A cuff that is appropriate for a child or adult with a thin arm.</items>
</term_definitions>
<term_definitions code="at1010">
<items id="text">Diastolic endpoint</items>
<items id="description">Record which Korotkoff sound is used for determining diastolic pressure using auscultative method.</items>
</term_definitions>
<term_definitions code="at1011">
<items id="text">Phase IV</items>
<items id="description">The fourth Korotkoff sound is identified as an abrupt muffling of sounds.</items>
</term_definitions>
<term_definitions code="at1012">
<items id="text">Phase V</items>
<items id="description">The fifth Korotkoff sound is identified by absence of sounds as the cuff pressure drops below the diastolic blood pressure.</items>
</term_definitions>
<term_definitions code="at1014">
<items id="text">Lying with tilt to left</items>
<items id="description">Lying flat with some lateral tilt, usually angled towards the left side. Commonly required in the last trimester of pregnancy to relieve aortocaval compression.</items>
</term_definitions>
<term_definitions code="at1018">
<items id="text">Infant</items>
<items id="description">A cuff used for infants.</items>
</term_definitions>
<term_definitions code="at1019">
<items id="text">Neonatal</items>
<items id="description">A cuff used for a neonate, assuming cuff is the appropriate size for maturity and birthweight of the neonate.</items>
</term_definitions>
<term_definitions code="at1020">
<items id="text">Right wrist</items>
<items id="description">The right wrist of the individual.</items>
</term_definitions>
<term_definitions code="at1021">
<items id="text">Left wrist</items>
<items id="description">The left wrist of the individual.</items>
</term_definitions>
<term_definitions code="at1025">
<items id="text">Device</items>
<items id="description">Details about sphygmomanometer or other device used to measure the blood pressure.</items>
</term_definitions>
<term_definitions code="at1026">
<items id="text">Right ankle</items>
<items id="description">The right ankle of the individual.</items>
</term_definitions>
<term_definitions code="at1030">
<items id="text">Exertion</items>
<items id="description">Details about physical activity undertaken at the time of blood pressure measurement.</items>
</term_definitions>
<term_definitions code="at1031">
<items id="text">Left ankle</items>
<items id="description">The left ankle of the individual.</items>
</term_definitions>
<term_definitions code="at1032">
<items id="text">Finger</items>
<items id="description">A finger of the individual.</items>
</term_definitions>
<term_definitions code="at1035">
<items id="text">Method</items>
<items id="description">Method of measurement of blood pressure.</items>
</term_definitions>
<term_definitions code="at1036">
<items id="text">Auscultation</items>
<items id="description">Method of measuring blood pressure externally, using a stethoscope and Korotkoff sounds.</items>
</term_definitions>
<term_definitions code="at1037">
<items id="text">Palpation</items>
<items id="description">Method of measuring blood pressure externally, using palpation (usually of the brachial or radial arteries).</items>
</term_definitions>
<term_definitions code="at1038">
<items id="text">Mean arterial pressure formula</items>
<items id="description">Formula used to calculate the Mean Arterial Pressure (if recorded in data).</items>
</term_definitions>
<term_definitions code="at1039">
<items id="text">Machine</items>
<items id="description">Method of measuring blood pressure externally, using a blood pressure machine.</items>
</term_definitions>
<term_definitions code="at1040">
<items id="text">Invasive</items>
<items id="description">Method of measuring blood pressure internally ie involving penetration of the skin and measuring inside blood vessels.</items>
</term_definitions>
<term_definitions code="at1042">
<items id="text">24 hour average</items>
<items id="description">Estimate of the average blood pressure over a 24 hour period.</items>
</term_definitions>
<term_definitions code="at1043">
<items id="text">Sleep status</items>
<items id="description">Sleep status - supports interpretation of 24 hour ambulatory blood pressure records.</items>
</term_definitions>
<term_definitions code="at1044">
<items id="text">Awake</items>
<items id="description">The individual is fully conscious.</items>
</term_definitions>
<term_definitions code="at1045">
<items id="text">Sleeping</items>
<items id="description">The individual is in the natural state of bodily rest.</items>
</term_definitions>
<term_definitions code="at1051">
<items id="text">Toe</items>
<items id="description">A toe of the individual.</items>
</term_definitions>
<term_definitions code="at1052">
<items id="text">Confounding factors</items>
<items id="description">Comment on and record other incidental factors that may be contributing to the blood pressure measurement. For example, level of anxiety or 'white coat syndrome'; pain or fever; changes in atmospheric pressure etc.</items>
</term_definitions>
<term_definitions code="at1053">
<items id="text">Intra-arterial</items>
<items id="description">Invasive measurement via transducer access line within an artery.</items>
</term_definitions>
<term_definitions code="at1054">
<items id="text">Systolic pressure formula</items>
<items id="description">Formula used to calculate the systolic pressure from from mean arterial pressure (if recorded in data).</items>
</term_definitions>
<term_definitions code="at1055">
<items id="text">Diastolic pressure formula</items>
<items id="description">Formula used to calculate the diastolic pressure from mean arterial pressure (if recorded in data).</items>
</term_definitions>
<term_definitions code="at1057">
<items id="text">Structured measurement location</items>
<items id="description">Structured anatomical location of where the measurement was taken.</items>
</term_definitions>
<term_definitions code="at1058">
<items id="text">Extension</items>
<items id="description">Additional information required to capture local context or to align with other reference models/formalisms.</items>
<items id="comment">For example: Local hospital departmental infomation or additional metadata to align with FHIR or CIMI equivalents.</items>
</term_definitions>
<term_definitions code="at1059">
<items id="text">Clinical interpretation</items>
<items id="description">Single word, phrase or brief description that represents the clinical meaning and significance of the blood pressure measurement.</items>
</term_definitions>
<term_definitions code="at1056">
<items id="text">Dorsum of foot</items>
<items id="description">The individual's dorsum of the foot.</items>
</term_definitions>
<term_definitions code="at0000">
<items id="text">Blood pressure</items>
<items id="description">The local measurement of arterial blood pressure which is a surrogate for arterial pressure in the systemic circulation.</items>
<items id="comment">Most commonly, use of the term 'blood pressure' refers to measurement of brachial artery pressure in the upper arm.</items>
</term_definitions>
<term_definitions code="mm[Hg]">
<items id="text">mmHg</items>
<items id="description">millimeters of mercury</items>
</term_definitions>
<term_bindings terminology="SNOMED-CT">
<items code="at0004">
<value>
<terminology_id>
<value>SNOMED-CT</value>
</terminology_id>
<code_string>[SNOMED-CT(2003)::271649006]</code_string>
</value>
</items>
<items code="at0005">
<value>
<terminology_id>
<value>SNOMED-CT</value>
</terminology_id>
<code_string>[SNOMED-CT(2003)::271650006]</code_string>
</value>
</items>
<items code="at0013">
<value>
<terminology_id>
<value>SNOMED-CT</value>
</terminology_id>
<code_string>[SNOMED-CT(2003)::246153002]</code_string>
</value>
</items>
<items code="at0000">
<value>
<terminology_id>
<value>SNOMED-CT</value>
</terminology_id>
<code_string>[SNOMED-CT(2003)::364090009]</code_string>
</value>
</items>
</term_bindings>
</children>
<cardinality>
<is_ordered>false</is_ordered>
<is_unique>false</is_unique>
<interval>
<lower_included>true</lower_included>
<upper_included>false</upper_included>
<lower_unbounded>false</lower_unbounded>
<upper_unbounded>true</upper_unbounded>
<lower>1</lower>
</interval>
</cardinality>
</attributes>
<archetype_id>
<value>openEHR-EHR-COMPOSITION.encounter.v1</value>
</archetype_id>
<template_id>
<value>Blood Pressure</value>
</template_id>
<term_definitions code="at0001">
<items id="text">Tree</items>
<items id="description">@ internal @</items>
</term_definitions>
<term_definitions code="at0002">
<items id="text">Extension</items>
<items id="description">Additional information required to extend the model with local content or to align with other reference models or formalisms.</items>
<items id="comment">For example: local information requirements; or additional metadata to align with FHIR.</items>
</term_definitions>
<term_definitions code="at0000">
<items id="text">Blood Pressure</items>
<items id="description">Interaction, contact or care event between a subject of care and healthcare provider(s).</items>
</term_definitions>
</definition>
</template>'The response should be an echo of the uploaded template with a 201 Created status, if the template didn't exist, or a 409 Conflict status if the template already exists.
Commit Clinical Data
Let's generate a valid openEHR document sample from the Template we just uploaded. For that you can use the openEHR Toolkit.
- Create a free account
- Login
- Go to Templates
- Upload our Blood Pressure template
- Click on Canonical Instance Generator
- Click on Generate
- Copy the generated data in JSON format
The generated instance looks like this:
{
"_type": "COMPOSITION",
"name": {
"_type": "DV_TEXT",
"value": "Blood Pressure"
},
"archetype_details": {
"archetype_id": {
"value": "openEHR-EHR-COMPOSITION.encounter.v1"
},
"template_id": {
"value": "Blood Pressure"
},
"rm_version": "1.0.2"
},
"archetype_node_id": "openEHR-EHR-COMPOSITION.encounter.v1",
"language": {
"terminology_id": {
"_type": "TERMINOLOGY_ID",
"value": "ISO_639-1"
},
"code_string": "en"
},
"territory": {
"terminology_id": {
"_type": "TERMINOLOGY_ID",
"value": "ISO_3166-1"
},
"code_string": "UY"
},
"category": {
"value": "event",
"defining_code": {
"terminology_id": {
"_type": "TERMINOLOGY_ID",
"value": "openehr"
},
"code_string": "433"
}
},
"composer": {
"_type": "PARTY_IDENTIFIED",
"external_ref": {
"id": {
"_type": "HIER_OBJECT_ID",
"value": "799f4736-3f3f-4223-a5d5-029c48d44c19"
},
"namespace": "DEMOGRAPHIC",
"type": "PERSON"
},
"name": "Dr. Yamamoto"
},
"context": {
"start_time": {
"value": "2025-12-22T06:04:49.335Z"
},
"setting": {
"value": "home",
"defining_code": {
"terminology_id": {
"_type": "TERMINOLOGY_ID",
"value": "openehr"
},
"code_string": "225"
}
},
"other_context": {
"_type": "ITEM_TREE",
"name": {
"_type": "DV_TEXT",
"value": "Tree"
},
"archetype_node_id": "at0001",
"items": []
}
},
"content": [
{
"_type": "OBSERVATION",
"name": {
"_type": "DV_TEXT",
"value": "Blood pressure"
},
"archetype_details": {
"archetype_id": {
"value": "openEHR-EHR-OBSERVATION.blood_pressure.v2"
},
"template_id": {
"value": "Blood Pressure"
},
"rm_version": "1.0.2"
},
"archetype_node_id": "openEHR-EHR-OBSERVATION.blood_pressure.v2",
"language": {
"terminology_id": {
"_type": "TERMINOLOGY_ID",
"value": "ISO_639-1"
},
"code_string": "en"
},
"encoding": {
"terminology_id": {
"_type": "TERMINOLOGY_ID",
"value": "IANA_character-sets"
},
"code_string": "UTF-8"
},
"subject": {
"_type": "PARTY_SELF"
},
"protocol": {
"_type": "ITEM_TREE",
"name": {
"_type": "DV_TEXT",
"value": "Tree"
},
"archetype_node_id": "at0011",
"items": []
},
"data": {
"_type": "HISTORY",
"name": {
"_type": "DV_TEXT",
"value": "History"
},
"archetype_node_id": "at0001",
"origin": {
"value": "2025-12-22T06:04:49.448Z"
},
"events": [
{
"_type": "POINT_EVENT",
"name": {
"_type": "DV_TEXT",
"value": "Any event"
},
"archetype_node_id": "at0006",
"time": {
"value": "2025-12-22T06:04:49.450Z"
},
"data": {
"_type": "ITEM_TREE",
"name": {
"_type": "DV_TEXT",
"value": "blood pressure"
},
"archetype_node_id": "at0003",
"items": [
{
"_type": "ELEMENT",
"name": {
"_type": "DV_TEXT",
"value": "Systolic"
},
"archetype_node_id": "at0004",
"value": {
"_type": "DV_QUANTITY",
"magnitude": 549.7,
"units": "mm[Hg]"
}
},
{
"_type": "ELEMENT",
"name": {
"_type": "DV_TEXT",
"value": "Diastolic"
},
"archetype_node_id": "at0005",
"value": {
"_type": "DV_QUANTITY",
"magnitude": 903.9,
"units": "mm[Hg]"
}
},
{
"_type": "ELEMENT",
"name": {
"_type": "DV_TEXT",
"value": "Pulse pressure"
},
"archetype_node_id": "at1007",
"value": {
"_type": "DV_QUANTITY",
"magnitude": 55.2,
"units": "mm[Hg]"
}
},
{
"_type": "ELEMENT",
"name": {
"_type": "DV_TEXT",
"value": "Clinical interpretation"
},
"archetype_node_id": "at1059",
"value": {
"_type": "DV_TEXT",
"value": "TxzpdBIAoDOwU....xaOGaOapJwY.nHpGKbgZ"
}
},
{
"_type": "ELEMENT",
"name": {
"_type": "DV_TEXT",
"value": "Comment"
},
"archetype_node_id": "at0033",
"value": {
"_type": "DV_TEXT",
"value": "loRZkegxIgvHGmftKq ZOnswS...zJtEGsMTgytYXCpQHM"
}
}
]
},
"state": {
"_type": "ITEM_TREE",
"name": {
"_type": "DV_TEXT",
"value": "state structure"
},
"archetype_node_id": "at0007",
"items": [
{
"_type": "ELEMENT",
"name": {
"_type": "DV_TEXT",
"value": "Position"
},
"archetype_node_id": "at0008",
"value": {
"_type": "DV_CODED_TEXT",
"value": "Standing",
"defining_code": {
"terminology_id": {
"_type": "TERMINOLOGY_ID",
"value": "local"
},
"code_string": "at1000"
}
}
}
]
}
}
]
}
}
]
}Commit Data
Here, we use the ehr_id (d5384e04-eff4-45a4-94c8-c98ec17cdf42) we got from the Create EHR step.
curl --request POST \
--url https://your-instance.atomik.app/api/v1/ehr/d5384e04-eff4-45a4-94c8-c98ec17cdf42/composition \
--header 'Authorization: Bearer eyJhbGciOiJIUzI1Ni...Lc2ZtnR7ijjgma2qtLg' \
--header 'Content-Type: application/json' \
--header 'Prefer: return=minimal' \
--header 'openEHR-AUDIT_DETAILS.committer: name="John Doe", external_ref.id="BC8132EA-8F4A-11E7-BB31-BE2E44B06B34", external_ref.namespace="demographic", external_ref.type="PERSON"' \
--header 'openEHR-AUDIT_DETAILS.description: value="A new composition description"' \
--data '__GENERATED_DOCUMENT_GOES_HERE__'✅ If you get a 204 No Content response, then it's a success! You've stored your first clinical record in Atomik.
Note in the response headers you will get some important information:
- ETag: e678c270-1074-450b-a8de-0de20777feb4::CABOLABS_ATOMIK_SERVER::1
- Location: /e678c270-1074-450b-a8de-0de20777feb4::CABOLABS_ATOMIK_SERVER::1
The ETag is the identifier of the document, and the Location is the relative location of that document from the current URL (https://your-instance.atomik.app/api/v1/ehr/d5384e04-eff4-45a4-94c8-c98ec17cdf42/composition)
Step 4: Query and Retrieve Data
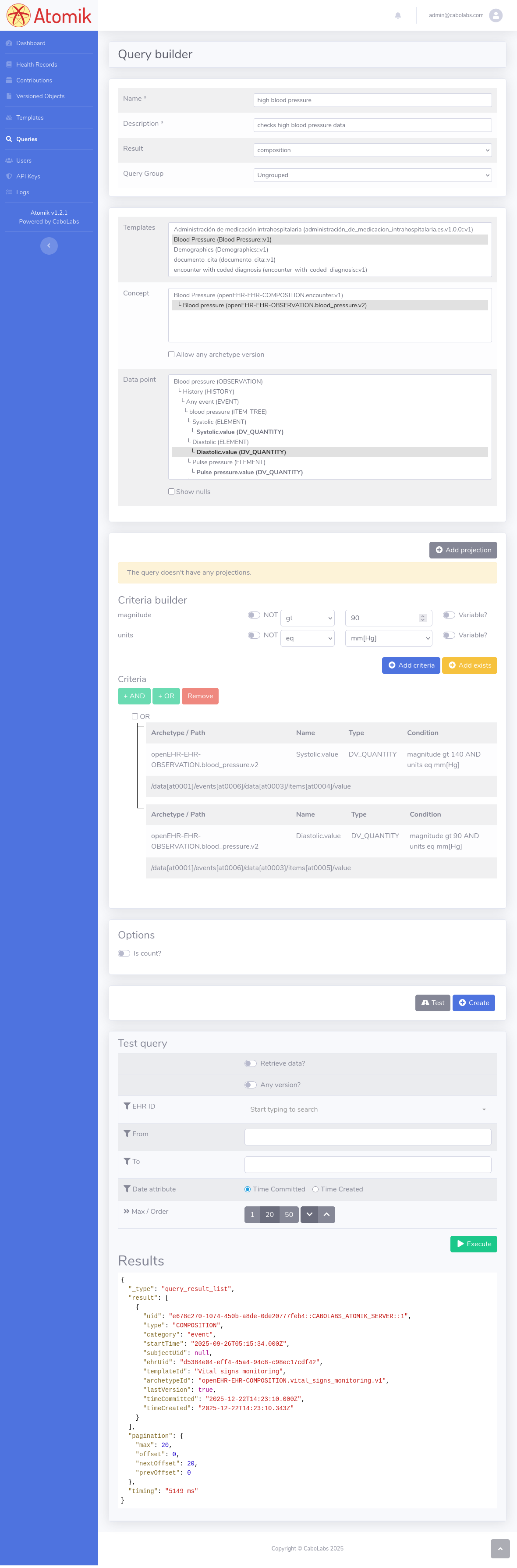
These are the steps to create a query:
- Login to Atomik's Web Console: https://your-instance.atomik.app
- Go to the Queries section
- Click on the + button
- Enter a name and description for the query
- Select the type of result "composition"
- From the template list, select the "Blood Pressure" template
- In the Concept list select the OBSERVATION.blood_pressure archetype
- In the Data point selector, select the "systolic.value" data point
- In the Criteria Builder section, add a condition for "magnitude > 140", click on Add criteria
- Now select the "diastolic.value" from the Data point selector
- Again in the Criteria Builder, add a condition for "magnitude > 90", click on Add criteria
- In the Criteria section, select both conditions and click on OR, that will add a complex OR condition
- Click on Test
- Click on Execute (select Retrieve Data if you want the full document to be retrieved in the result)
- If there is some document that complies with the conditions, it will be shown in the query results
- Click on Create, that will store the query to be executed later
- In the Query Details, you can find the identifier of the query, that will be used from the API to execute the query
Execute the query from the API
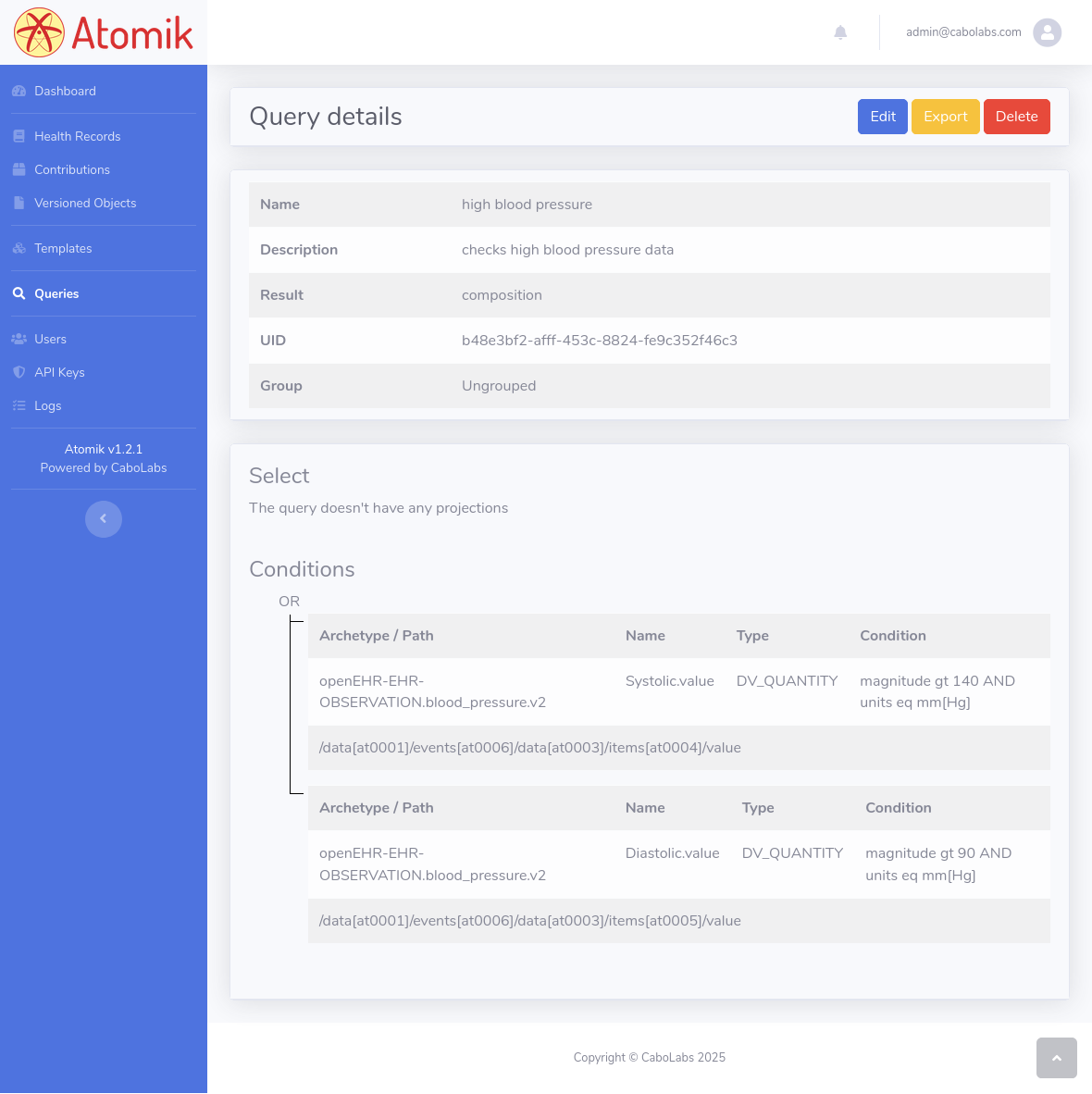
Here, we use the query_uid (b48e3bf2-afff-453c-8824-fe9c352f46c3) we got from the Query Creation flow described above. Check the Query Builder screenshot at the bottom of this guide as a reference.
curl --request GET \
--url 'https://your-instance.atomik.app/api/v1/query/b48e3bf2-afff-453c-8824-fe9c352f46c3/execute?retrieveData=true' \
--header 'Authorization: Bearer eyJhbGciOiJIUzI1Ni...Lc2ZtnR7ijjgma2qtLg' \
Query result:
Note the query result contains item metadata for each result item, pagination and timing elements.
{
"_type": "query_result_list",
"result": [
{
"meta": {
"ehr_uid": "d5384e04-eff4-45a4-94c8-c98ec17cdf42",
"subject_id": null,
"time_created": "2025-12-22T14:23:10Z"
},
"locatable": {
"_type": "COMPOSITION",
"name": {
"_type": "DV_TEXT",
"value": "Blood Pressure"
},
"archetype_details": {
"archetype_id": {
"value": "openEHR-EHR-COMPOSITION.encounter.v1"
},
"template_id": {
"value": "Blood Pressure"
},
"rm_version": "1.0.2"
},
"archetype_node_id": "openEHR-EHR-COMPOSITION.encounter.v1",
"language": {
"terminology_id": {
"_type": "TERMINOLOGY_ID",
"value": "ISO_639-1"
},
"code_string": "en"
},
"territory": {
"terminology_id": {
"_type": "TERMINOLOGY_ID",
"value": "ISO_3166-1"
},
"code_string": "UY"
},
"category": {
"value": "event",
"defining_code": {
"terminology_id": {
"_type": "TERMINOLOGY_ID",
"value": "openehr"
},
"code_string": "433"
}
},
"composer": {
"_type": "PARTY_IDENTIFIED",
"external_ref": {
"id": {
"_type": "HIER_OBJECT_ID",
"value": "799f4736-3f3f-4223-a5d5-029c48d44c19"
},
"namespace": "DEMOGRAPHIC",
"type": "PERSON"
},
"name": "Dr. Yamamoto"
},
"context": {
"start_time": {
"value": "2025-12-22T06:04:49.335Z"
},
"setting": {
"value": "home",
"defining_code": {
"terminology_id": {
"_type": "TERMINOLOGY_ID",
"value": "openehr"
},
"code_string": "225"
}
},
"other_context": {
"_type": "ITEM_TREE",
"name": {
"_type": "DV_TEXT",
"value": "Tree"
},
"archetype_node_id": "at0001",
"items": []
}
},
"content": [
{
"_type": "OBSERVATION",
"name": {
"_type": "DV_TEXT",
"value": "Blood pressure"
},
"archetype_details": {
"archetype_id": {
"value": "openEHR-EHR-OBSERVATION.blood_pressure.v2"
},
"template_id": {
"value": "Blood Pressure"
},
"rm_version": "1.0.2"
},
"archetype_node_id": "openEHR-EHR-OBSERVATION.blood_pressure.v2",
"language": {
"terminology_id": {
"_type": "TERMINOLOGY_ID",
"value": "ISO_639-1"
},
"code_string": "en"
},
"encoding": {
"terminology_id": {
"_type": "TERMINOLOGY_ID",
"value": "IANA_character-sets"
},
"code_string": "UTF-8"
},
"subject": {
"_type": "PARTY_SELF"
},
"protocol": {
"_type": "ITEM_TREE",
"name": {
"_type": "DV_TEXT",
"value": "Tree"
},
"archetype_node_id": "at0011",
"items": []
},
"data": {
"_type": "HISTORY",
"name": {
"_type": "DV_TEXT",
"value": "History"
},
"archetype_node_id": "at0001",
"origin": {
"value": "2025-12-22T06:04:49.448Z"
},
"events": [
{
"_type": "POINT_EVENT",
"name": {
"_type": "DV_TEXT",
"value": "Any event"
},
"archetype_node_id": "at0006",
"time": {
"value": "2025-12-22T06:04:49.450Z"
},
"data": {
"_type": "ITEM_TREE",
"name": {
"_type": "DV_TEXT",
"value": "blood pressure"
},
"archetype_node_id": "at0003",
"items": [
{
"_type": "ELEMENT",
"name": {
"_type": "DV_TEXT",
"value": "Systolic"
},
"archetype_node_id": "at0004",
"value": {
"_type": "DV_QUANTITY",
"magnitude": 549.7,
"units": "mm[Hg]"
}
},
{
"_type": "ELEMENT",
"name": {
"_type": "DV_TEXT",
"value": "Diastolic"
},
"archetype_node_id": "at0005",
"value": {
"_type": "DV_QUANTITY",
"magnitude": 903.9,
"units": "mm[Hg]"
}
},
{
"_type": "ELEMENT",
"name": {
"_type": "DV_TEXT",
"value": "Pulse pressure"
},
"archetype_node_id": "at1007",
"value": {
"_type": "DV_QUANTITY",
"magnitude": 55.2,
"units": "mm[Hg]"
}
},
{
"_type": "ELEMENT",
"name": {
"_type": "DV_TEXT",
"value": "Clinical interpretation"
},
"archetype_node_id": "at1059",
"value": {
"_type": "DV_TEXT",
"value": "TxzpdBIAoDOwU....xaOGaOapJwY.nHpGKbgZ"
}
},
{
"_type": "ELEMENT",
"name": {
"_type": "DV_TEXT",
"value": "Comment"
},
"archetype_node_id": "at0033",
"value": {
"_type": "DV_TEXT",
"value": "loRZkegxIgvHGmftKq ZOnswS...zJtEGsMTgytYXCpQHM"
}
}
]
},
"state": {
"_type": "ITEM_TREE",
"name": {
"_type": "DV_TEXT",
"value": "state structure"
},
"archetype_node_id": "at0007",
"items": [
{
"_type": "ELEMENT",
"name": {
"_type": "DV_TEXT",
"value": "Position"
},
"archetype_node_id": "at0008",
"value": {
"_type": "DV_CODED_TEXT",
"value": "Standing",
"defining_code": {
"terminology_id": {
"_type": "TERMINOLOGY_ID",
"value": "local"
},
"code_string": "at1000"
}
}
}
]
}
}
]
}
}
]
}
}
],
"pagination": {
"max": 20,
"offset": 0,
"nextOffset": 20,
"prevOffset": 0
},
"timing": "508 ms"
}✅ You've successfully queried your data!
What's next?
Explore core concepts
- openEHR Basics - understand the foundation of Atomik's data model and design principles
- Templates - learn how Atomik manages data structure definitions
- Queries - master Atomik's querying engine for data retrieval
Common use cases
- Monitoring & Wearables - Stream data from IoT devices
- Clinical Decision Support - Build intelligent health applications
- Shared Health Records - Create interoperable patient record
Deep dives
- API Reference - Complete endpoint documentation
- Managing Data Models - Create and configure templates
- Data Versioning - Understand Atomik's audit capabilities
- Electronic Signature - Understand how openEHR's attestations are handled in Atomik
Integrations
- Integration Basics -
- Loading and Retrieving Data -
- Security Considerations - Implement proper security measures for each scenario
Need help?
Ready to build something amazing? You now have everything you need to start integrating Atomik into your application. Happy coding! 🚀
Screenshots with all the steps.